We have all heard about DNA. It is passed from parents to their children and determines whether you get blue or brown eyes, what is used to identify criminals and stuff. The genetic information that all living beings have and makes them be what they are. In general, we understand, with differing depth, that DNA (genes) is what gives us our individuality.
Our physiognomy, innate abilities and even part of our character are mostly defined by our DNA. It is what makes us different to other people and unique. But what happens in the case of identical twins? They are perfect genetic clones; can they really be different individuals? As much as they share the same genes and set off from the same basis, so to say, two identical twins will never look really identical. Food, weather, life experiences, etc. also play a crucial role in the development of any living being and they forge their individuality. Therefore, the famous deoxyribonucleic acid (whence the acronym, DNA) is not the only thing that defines an organism, and even two genetically identical clones can develop differently.
Although all of this is fairly well assimilated when it comes to humans, we do not apply it the same way when we refer to unicellular organisms like yeasts or bacteria. These often replicate asexually, meaning that from one single parental cell, one can obtain a whole army of clones carrying the same DNA. It is widely assumed that all the offspring must be and behave in the same way, since they have the same genes. In fact, when examining a group of cells, average values of their characteristics are always measured, like average size, mean resistance to a drug, average production of a given substance, etc. These values represent a cell population, but within such group every individual presents specific values, sometimes considerably far from the official average. These differences are often due to the fact that each cell is exposed to subtle variations in temperature, nutrients or oxygen. But it turns out that clone cells also display differences even when they are all in an identical environment.
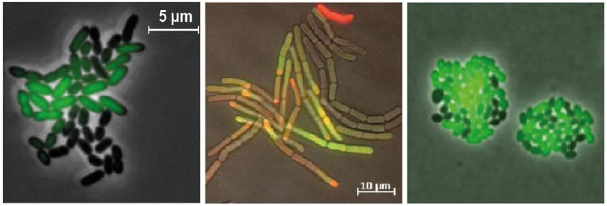
Bacterial clones with different shapes and fluorescence.
Why do these differences occur among theoretical perfect clones if there are no variations in the outer medium? One would think that with cells being the most basic forms of live, replicating them without mistake should be straightforward. The reality is that a cell in itself has multiple levels of complexity, millions of molecules and thus many points where things can go wrong. Every cell process is susceptible to stochasticity (a sophisticated synonym for chance). Despite the strict control exerted by DNA, little random variations in the cellular components occur, thus generating “noise”, the genetic information being distorted among clone cells. Besides, this noise is propagated within a cell: every minimal imbalance concerning a process or molecule can in turn affect others and eventually alter cell size, shape or resistance to toxic compounds, among other features. Therefore, even when sharing the same genes two clones can become pretty different in several aspects due to those “noisy errors” caused by chance.
But are these mere errors? Curiously enough, it has been observed that the level of randomness must be somehow regulated, since some cell processes display more noise than others. In general, basic cell functions (like core metabolism or cell division) happen to be quite robust, there is very little variation between clones. However, there are other processes, mainly secondary, for which two genetically identical cells can vary greatly. What does this imply? Essentially, that the level of noise must play some role, given the fact that it seems to be controlled for every specific process. Moreover, the level of variation depends on the kind of cells in question, and it can evolve through natural selection (and other forces) towards a higher or lower noise.
There are several explanations for the existence of such a controlled noise, but the most straightforward considers that this ability of a cell community to produce multiple profiles from the same DNA constitutes an adaptive strategy, known as bet-hedging. A cell population seeks above anything to secure its own survival, and for that, it bets on producing different individuals (although genetically identical). In that way, when external conditions suddenly change there will be at least a few cells that will survive and replicate to maintain the line. If all individuals were the same, any abrupt alteration in the environment could completely erase the whole population and their DNA would become extinct, for all the individuals would be equally sensitive to that change. Do not tell me these little cells are not clever. And they seemed so simple!
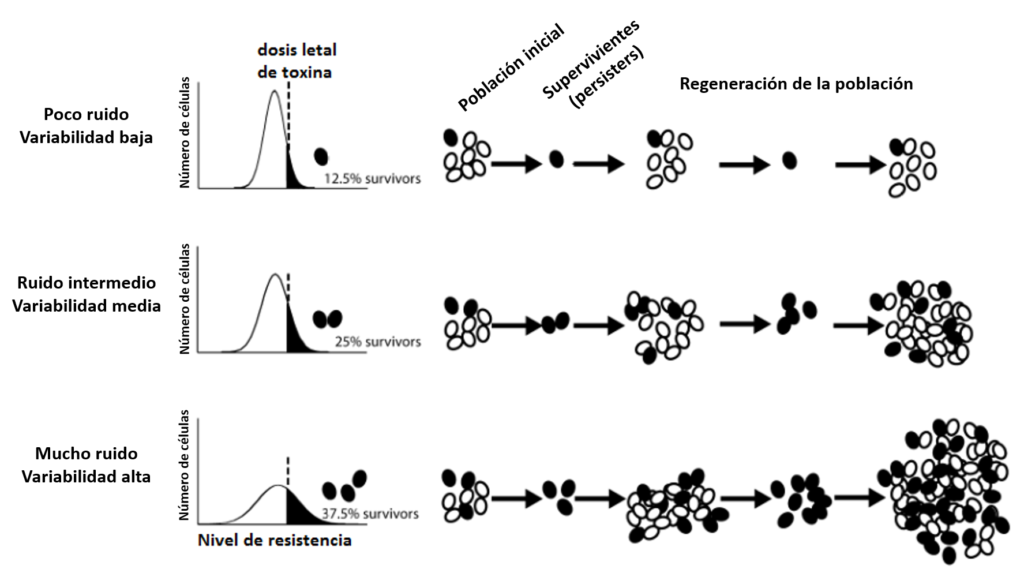
Bet-hedging: the more variability in a population, the higher the chances of surviving after an abrupt alteration in the environment.
We might be happy about a tribe of harmless bacteria being able to survive a draught in the middle of the field. However, if I get an infection and some of the causing microorganisms do not respond to the treatment because of their higher resistance, then that bet-hedging thing becomes less fun. This is currently the biggest challenge that genetic noise poses, which has gone unnoticed so far; infections caused by bacteria or fungi, as well as tumours, do not always react to therapies because of the presence of a few highly resistant clones (persisters). When most of the population dies, these individuals survive, and when the treatment is interrupted (and it seems we have won) they divide and proliferate again, renewing the infection or cancer. The same happens with plant pathogens, which can ruin whole crops, and with water- or food-spoilage microbes. That is why it is necessary to gain a better understanding of this phenomenon and assume that every cell is unique despite being genetically identical to others, and it has the potential to ensure the survival of its community. Thanks to the use of sophisticated (that is, expensive) techniques that allow simultaneous analysis of thousands of cells individually (like flow cytometry or microfluidics) we are getting closer to understanding and handling those variations that make clones so different despite being genetically identical.
More info and references:
Ackermann, M. (2015). A functional perspective on phenotypic heterogeneity in microorganisms. Nature Reviews Microbiology, 13, 497-508.
Binder, D., Drepper, T., Jaeger, K.-E., Delvigne, F., Wiechert, W., Kohlheyer, D., & Grünberger, A. (2017). Homogenizing bacterial cell factories: Analysis and engineering of phenotypic heterogeneity. Metabolic Engineering, 42, 145–156.
Holland, S.L., Reader, T., Dyer, P.S. & Avery, S.V. (2013). Phenotypic heterogeneity is a selected trait in natural yeast populations subject to environmental stress. Environmental Microbiology 16(6), 1729–1740.
By Elena Moreno Martínez, PhD student at University of Nottingham. SRUK Midland constituency.